How analyze individual spectral lines#
Many of the tools in SpecpolFlow can be applied to a single line in an observed spectrum. In this example we analyze a few emission lines in the spectrum of a T Tauri star.
First, import SpecpolFlow and some packages that will help with data visualization.
import specpolFlow as pol
import pandas as pd
import matplotlib.pyplot as plt
1. Create individual line file#
In this tutorial, we use the UPENA normalized spectrum (IndividualLine_tutorialfiles/1423137pn.s) of the active cool star BP Tau.
We first load in the spectrum.
spec = pol.read_spectrum("IndividualLine_tutorialfiles/1423137pn.s")
To create extract an individual line from the spectrum, we have to define the transition wavelength lambda0 and a range with respect to the line center. In the example below, we select the HeI line at 667.815 nm in a 0.6 nm window.
# Create individual line profile
lambda0 = 667.815 # He i 667.815 nm
geff = 1.0 # He i 667.815 nm, used later for calculating Bz
lwidth = 0.3
prof = spec.individual_line(lambda0 = lambda0, lwidth = lwidth)
The individual_line function returns prof, which is really an LSD profile object, and inherits all the attributes from the LSD class. Therefore, you can use all the functionality discussed in the LSD Class Tutorial (i.e., one can normalize, slice, or shift the line profile).
Below, we visualize the individual line using the plot class function.
fig, axes = prof.plot()
axes[-1].axhline(1, color ='k', ls = '--')
<matplotlib.lines.Line2D at 0x7f040b391a50>
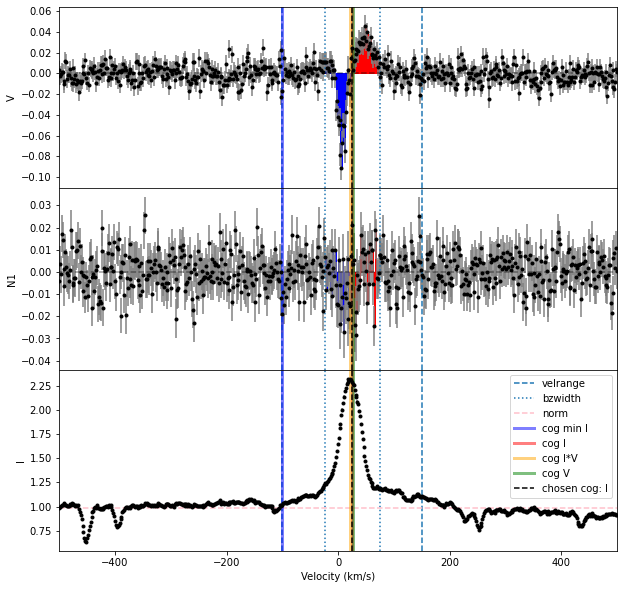
2. Calculate Bz#
To calculate the longitudinal field (Bz) of the individual line, we can use the calc_bz function of the LSD class.
# Define the limits for the continuum normalization
cmin = -50.0; cmax = 80.0
# Compute Bz in a 45 km/s window
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = [cmin, cmax], bzwidth = 45.0,
geff = geff, lambda0 = lambda0,
plot = True)
using AUTO method for the normalization
using the median of the continuum outside of the line
We can then display the results using the pandas dataframe. For this particular line, we find Bz \(= 1.2 \pm 0.1\) kG.
Bz_df = pd.DataFrame(data = [Bz])
# Simple display of the pandas dataframe
Bz_df.style
| V bz (G) | V bz sig (G) | V FAP | N1 bz (G) | N1 bz sig (G) | N1 FAP | N2 bz (G) | N2 bz sig (G) | N2 FAP | norm_method | norm | cog_method | cog | int. range start | int. range end | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1191.652271 | 130.184898 | 0.000000 | -34.686996 | 130.041356 | 0.564259 | 0.000000 | 0.000000 | 0.000000 | auto | 0.931285 | I | 21.173209 | -23.826791 | 66.173209 |
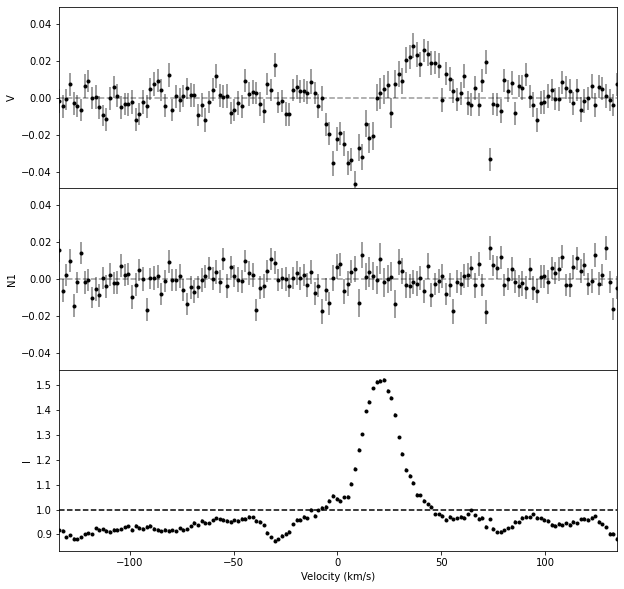
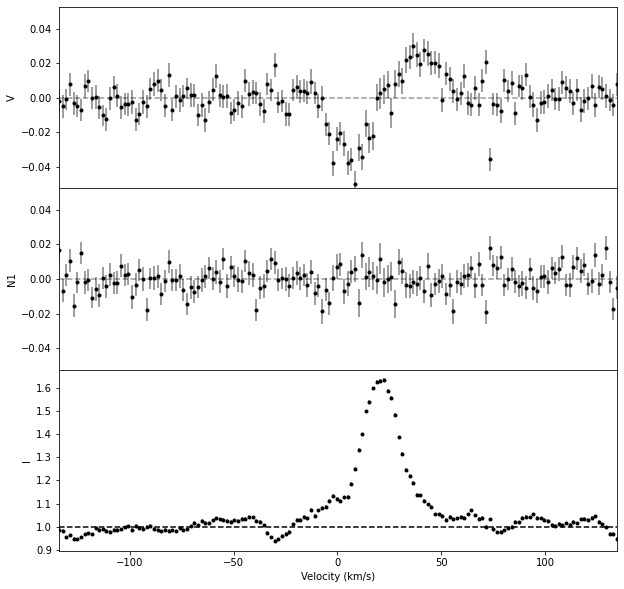
3. Normalize the individual line#
In the figure above, we see that calc_bz improved the continuum normalization, taking the median of the continuum outside of the line.
Therefore, we propagate this normalization in the prof object before saving the individual line profile into a file.
# Renormalize prof using the continuum value obtained by calc_bz
prof_norm = prof.norm(Bz['norm'])
# Display the profile
fig, axes = prof_norm.plot()
axes[-1].axhline(1, color = 'k', ls = '--')
<matplotlib.lines.Line2D at 0x7f0401fed2a0>
4. Saving individual lines#
Finally, you can save the individual line into a file using the save function, shown below. This file will have a text format like an LSD profile.
prof_norm.save("Output/1423137.Helium")
5. Dealing with order overlaps#
Now, let’s do the same for the Halpha line. However, there is a complication, the Halpha line is split across two orders in this echelle spectrum.
# reading the spectrum
spec = pol.read_spectrum("IndividualLine_tutorialfiles/1423137pn.s")
# defining the line parameters
lambda0 = 656.281 ; geff = 1.0 # Halpha
lwidth = 700. * lambda0 / 2.99e5 #convert a 700 km/s width into a width in nm
# getting the profile for that line
prof = spec.individual_line(lambda0 = lambda0, lwidth = lwidth)
# Define the limits for the continuum normalization
# Compute Bz in a 200 km/s window around the cog
cogRange = [-400., 400.]
intRange = 200.
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = True)
# Create a dataframe to record the result of the different methods in this section
d = {'Merge type':'None'}
d.update(Bz)
df_results = pd.DataFrame(data = [d])
df_results.style
using AUTO method for the normalization
using the median of the continuum outside of the line
/tmp/ipykernel_980312/871814974.py:13: UserWarning:
The velocity array is not monotonically increasing.
There might be an order overlap in the region of the observation used.
calc_bz will sort the LSD profile in velocity order.
Make sure this is what you want -- merge orders before running calc_bz()!
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
| Merge type | V bz (G) | V bz sig (G) | V FAP | N1 bz (G) | N1 bz sig (G) | N1 FAP | N2 bz (G) | N2 bz sig (G) | N2 FAP | norm_method | norm | cog_method | cog | int. range start | int. range end | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | 72.964936 | 17.843167 | 0.000000 | 7.405907 | 17.843162 | 0.435065 | 0.000000 | 0.000000 | 0.000000 | auto | 1.101100 | I | 9.725353 | -190.274647 | 209.725353 |
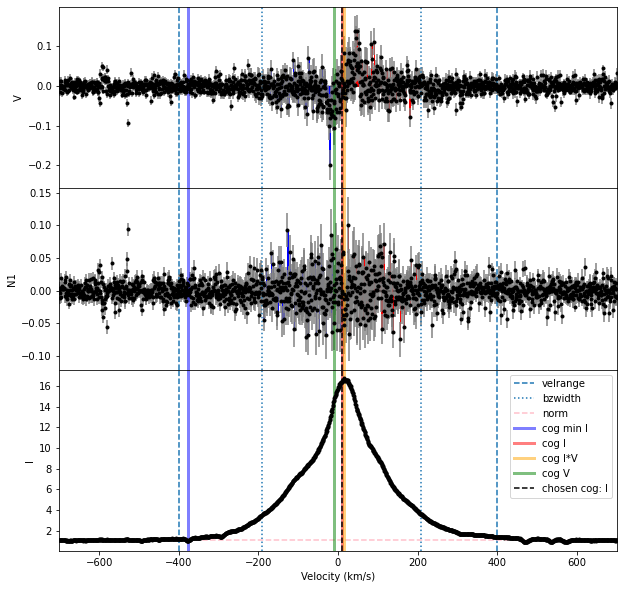
The calc_bz function gave us a warning about there probably being different spectral orders here. And the plot of results is relatively messy.
5.1 Option 1: Selecting only one order#
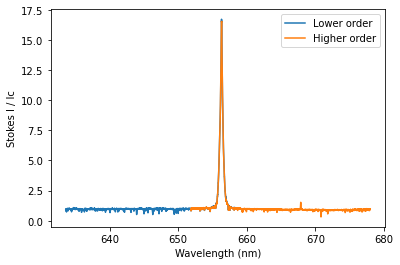
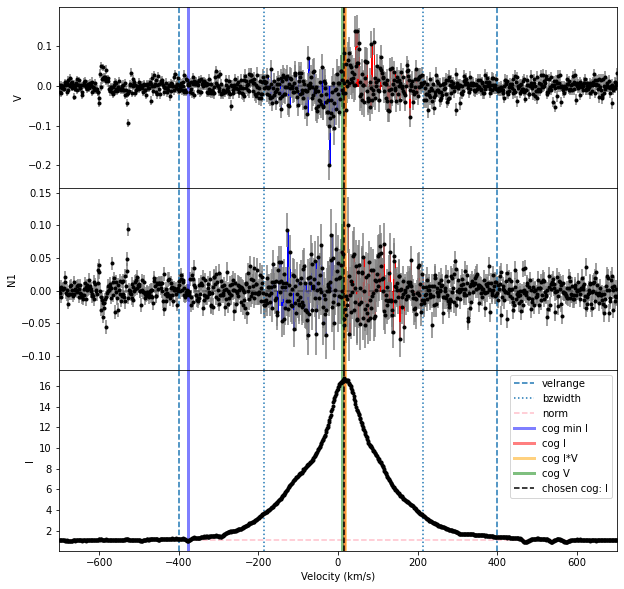
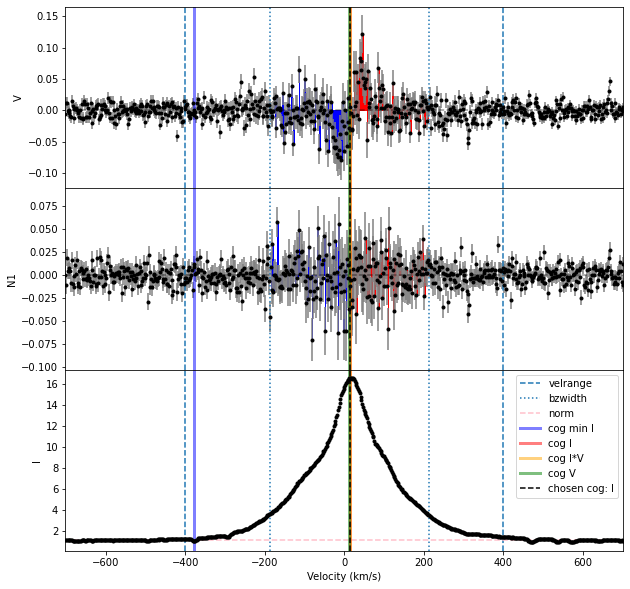
The simplest solution to this problem is to only use one spectral order for the Bz calculation. In the case of this Halpha line, both orders include the center of the line that we are interested in. So we can calculate Bz from the two orders separately.
To get the spectral orders that include a wavelength, or range of wavelengths, we can use the get_orders_in_range function of the Spectrum class. This returns a list containing Spectrum objects for each order.
## Selecting only one order
orders = spec.get_orders_in_range(lambda0)
print('There are {} orders with Halpha in them'.format(len(orders)))
# Illustration of the order overlap
fig, ax = plt.subplots(1,1)
ax.plot(orders[0].wl, orders[0].specI, label = 'Lower order')
ax.plot(orders[1].wl, orders[1].specI, label = 'Higher order')
ax.set_xlabel('Wavelength (nm)')
ax.set_ylabel('Stokes I / Ic')
ax.legend(loc = 0)
# loop over the orders
type = ['lower only', 'higher only']
i = 0
for ord in orders:
# get the profile for the line in this order
prof = ord.individual_line(lambda0 = lambda0, lwidth = lwidth)
# compute Bz, in the defined velocity window around the cog
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = True)
d = {'Merge type':type[i]}
d.update(Bz)
df = pd.DataFrame(data=[d])
df_results = pd.concat([df_results,df], ignore_index=True)
i += 1
# display the Bz result as a dataframe
df_results.style
There are 2 orders with Halpha in them
using AUTO method for the normalization
using the median of the continuum outside of the line
using AUTO method for the normalization
using the median of the continuum outside of the line
| Merge type | V bz (G) | V bz sig (G) | V FAP | N1 bz (G) | N1 bz sig (G) | N1 FAP | N2 bz (G) | N2 bz sig (G) | N2 FAP | norm_method | norm | cog_method | cog | int. range start | int. range end | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | 72.964936 | 17.843167 | 0.000000 | 7.405907 | 17.843162 | 0.435065 | 0.000000 | 0.000000 | 0.000000 | auto | 1.101100 | I | 9.725353 | -190.274647 | 209.725353 |
| 1 | lower only | 94.012588 | 25.428151 | 0.000003 | 5.209152 | 25.428140 | 0.221315 | 0.000000 | 0.000000 | 0.000000 | auto | 1.104800 | I | 14.197130 | -185.802870 | 214.197130 |
| 2 | higher only | 54.213318 | 21.662775 | 0.000007 | 3.435552 | 21.662771 | 0.803308 | 0.000000 | 0.000000 | 0.000000 | auto | 1.096300 | I | 13.943177 | -186.056823 | 213.943177 |
5.2 Option 2: merging the orders#
An alternative solution is to merge the spectral orders before calculating Bz. Combining orders is more complex, but it can optimize S/N, and it can be useful when the wavelength range of interest extends beyond the edge of an order.
We can combine spectral orders with the merge_orders function of the Spectrum class. This function provides two methods of combining orders 'trim' and 'coadd'. This function only really works properly if the continuum levels are consistent between orders.
In merge_orders with mode = 'trim', the function just uses the first order up to the midpoint of the region where the orders overlap, then it uses the second order beyond that midpoint. This approach is relatively simple and essentially throws away a bit of signal. But it is more robust against errors in continuum normalization (or spectrum extraction) at the edges of spectral orders, and these errors are relatively common.
In merge_orders with mode = 'coadd', the function essentially averages the orders, weighted by the uncertainties on individual pixels (specifically by \(1/\sigma^2\)). To do this it needs to interpolate in wavelength, and it uses the wavelengths from the first order, interpolating the second order onto that grid. This mode requires reliable uncertainties for the Stokes I spectrum. This approach has the advantage that it optimizes the S/N in the merged spectrum. But it is more vulnerable to continuum normalization errors at the edges of orders, since an error in either order will cause problems for the merged result.
Here we try out both methods to compare the results. In this case the normalization is pretty good in the region we care about, so the 'coadd' mode gives a more precise result.
## Merging using the trim method
merge = spec.merge_orders(mode = 'trim', midpoint = 0.5)
prof = merge.individual_line(lambda0 = lambda0, lwidth = lwidth)
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = True)
d = {'Merge type':'trim at mid-point'}
d.update(Bz)
df = pd.DataFrame(data = [d])
df_results = pd.concat([df_results,df], ignore_index=True)
## Merging using the coadd method
merge = spec.merge_orders(mode='coadd')
prof = merge.individual_line(lambda0 = lambda0, lwidth = lwidth)
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = True)
d = {'Merge type':'coadd'}
d.update(Bz)
df = pd.DataFrame(data = [d])
df_results = pd.concat([df_results,df], ignore_index = True)
# Display the Bz result as a dataframe
df_results.style
using AUTO method for the normalization
using the median of the continuum outside of the line
using AUTO method for the normalization
using the median of the continuum outside of the line
| Merge type | V bz (G) | V bz sig (G) | V FAP | N1 bz (G) | N1 bz sig (G) | N1 FAP | N2 bz (G) | N2 bz sig (G) | N2 FAP | norm_method | norm | cog_method | cog | int. range start | int. range end | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | 72.964936 | 17.843167 | 0.000000 | 7.405907 | 17.843162 | 0.435065 | 0.000000 | 0.000000 | 0.000000 | auto | 1.101100 | I | 9.725353 | -190.274647 | 209.725353 |
| 1 | lower only | 94.012588 | 25.428151 | 0.000003 | 5.209152 | 25.428140 | 0.221315 | 0.000000 | 0.000000 | 0.000000 | auto | 1.104800 | I | 14.197130 | -185.802870 | 214.197130 |
| 2 | higher only | 54.213318 | 21.662775 | 0.000007 | 3.435552 | 21.662771 | 0.803308 | 0.000000 | 0.000000 | 0.000000 | auto | 1.096300 | I | 13.943177 | -186.056823 | 213.943177 |
| 3 | trim at mid-point | 54.213318 | 21.662775 | 0.000007 | 3.435552 | 21.662771 | 0.803308 | 0.000000 | 0.000000 | 0.000000 | auto | 1.096300 | I | 13.943177 | -186.056823 | 213.943177 |
| 4 | coadd | 69.328145 | 15.158243 | 0.000000 | 5.479275 | 15.158239 | 0.999346 | 0.000000 | 0.000000 | 0.000000 | auto | 1.098296 | I | 14.040448 | -185.959552 | 214.040448 |
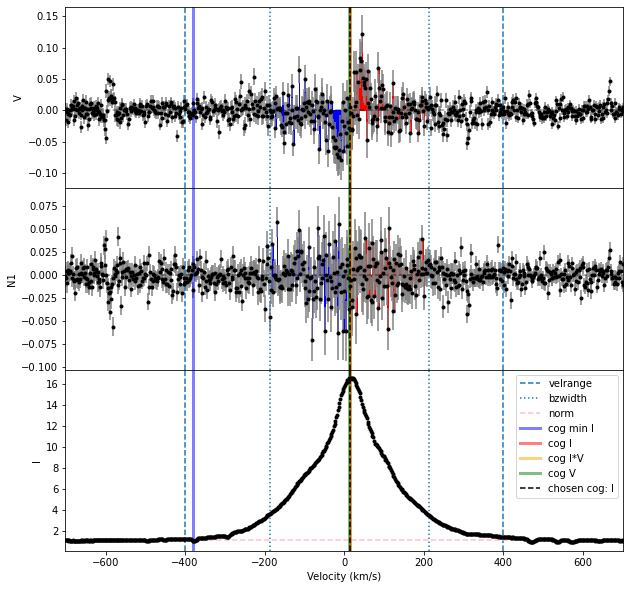
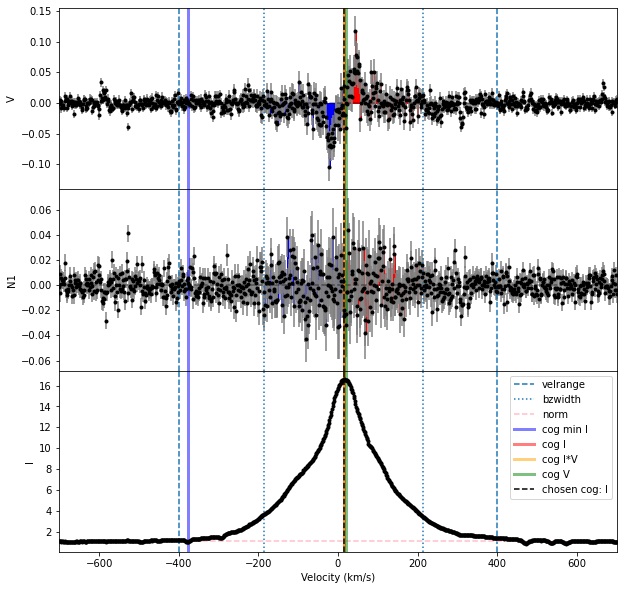
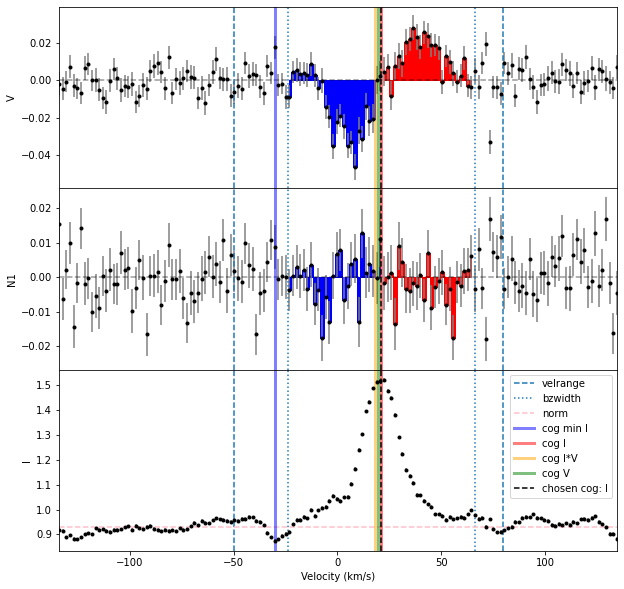
The Halpha line is somewhat complicated to analyze, in that it has a very broad emission component from regions not associated with the magnetic field. This makes the correct velocity range to use for integration in the Bz calculation ambiguous.
For a somewhat cleaner example that is still in a region where two spectral orders overlap, we can look at the He I 587.56 nm line.
# reading the spectrum
spec = pol.read_spectrum("IndividualLine_tutorialfiles/1423137pn.s")
# defining the line parameters
lambda0 = 587.56 ; geff = 1.0 # He I 587, really a multiplet (so geff is rather approximate)
lwidth = 500. * lambda0 / 2.99e5 #width in nm from width in km/s
# getting the profile for that line
prof = spec.individual_line(lambda0 = lambda0, lwidth = lwidth)
# define the limits for the continuum normalization
# compute Bz in a 50 km/s window around the cog
cogRange = [-100., 150.]
intRange = 50.
Bz = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = False)
# creating a dataframe to record the result of the different methods in this section
d = {'Merge type':'None'}
d.update(Bz)
df_results = pd.DataFrame(data = [d])
orders = spec.get_orders_in_range(lambda0)
# loop over the individual orders
for ord, type in zip(orders,['lower only', 'higher only']):
# get the profile for the line in this order
prof = ord.individual_line(lambda0 = lambda0, lwidth = lwidth)
Bz = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = False)
d = {'Merge type':type}
d.update(Bz)
df = pd.DataFrame(data = [d])
df_results = pd.concat([df_results,df], ignore_index=True)
# merging using the trim method
merge = spec.merge_orders(mode = 'trim', midpoint = 0.5)
prof = merge.individual_line(lambda0 = lambda0, lwidth = lwidth)
Bz = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = False)
d = {'Merge type':'trim at mid-point'}
d.update(Bz)
df = pd.DataFrame(data = [d])
df_results = pd.concat([df_results,df], ignore_index=True)
# merging using the coadd method
merge = spec.merge_orders(mode = 'coadd')
prof = merge.individual_line(lambda0 = lambda0, lwidth = lwidth)
Bz, fig = prof.calc_bz(norm = 'auto', cog = 'I',
velrange = cogRange, bzwidth = intRange,
geff = geff, lambda0 = lambda0,
plot = True)
d = {'Merge type':'coadd'}
d.update(Bz)
df = pd.DataFrame(data = [d])
df_results = pd.concat([df_results,df], ignore_index=True)
# display the Bz result as a dataframe
df_results.style
using AUTO method for the normalization
using the median of the continuum outside of the line
using AUTO method for the normalization
using the median of the continuum outside of the line
using AUTO method for the normalization
using the median of the continuum outside of the line
using AUTO method for the normalization
using the median of the continuum outside of the line
using AUTO method for the normalization
using the median of the continuum outside of the line
/tmp/ipykernel_980312/2408171684.py:13: UserWarning:
The velocity array is not monotonically increasing.
There might be an order overlap in the region of the observation used.
calc_bz will sort the LSD profile in velocity order.
Make sure this is what you want -- merge orders before running calc_bz()!
Bz = prof.calc_bz(norm = 'auto', cog = 'I',
| Merge type | V bz (G) | V bz sig (G) | V FAP | N1 bz (G) | N1 bz sig (G) | N1 FAP | N2 bz (G) | N2 bz sig (G) | N2 FAP | norm_method | norm | cog_method | cog | int. range start | int. range end | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | None | 699.503139 | 66.521645 | 0.000000 | 49.584163 | 66.504701 | 0.000385 | 0.000000 | 0.000000 | 0.000000 | auto | 0.991090 | I | 18.941158 | -31.058842 | 68.941158 |
| 1 | lower only | 822.700767 | 91.691815 | 0.000000 | -17.652850 | 91.658895 | 0.031436 | 0.000000 | 0.000000 | 0.000000 | auto | 0.992395 | I | 25.706350 | -24.293650 | 75.706350 |
| 2 | higher only | 793.994679 | 90.964654 | 0.000000 | 220.357069 | 90.937349 | 0.002410 | 0.000000 | 0.000000 | 0.000000 | auto | 0.990170 | I | 25.578820 | -24.421180 | 75.578820 |
| 3 | trim at mid-point | 797.884180 | 91.419382 | 0.000000 | 221.513065 | 91.391673 | 0.002410 | 0.000000 | 0.000000 | 0.000000 | auto | 0.993520 | I | 25.587490 | -24.412510 | 75.587490 |
| 4 | coadd | 805.924582 | 60.001692 | 0.000000 | 116.774813 | 59.981432 | 0.097482 | 0.000000 | 0.000000 | 0.000000 | auto | 0.990281 | I | 25.660364 | -24.339636 | 75.660364 |